RF-Cloning.org Forum
Anything and everything cloning: Go...
You are not logged in.
- Topics: Active | Unanswered
Pages: 1
#1 2014-10-15 13:14:19
- Dungeonlord
- Member
- Registered: 2014-10-13
- Posts: 2
Is phosphorilation needed?
Hi everyone
I have a very simple question. I was thinking of using the exponential Megaprimer PCR described by A. Ulrich 2012. In this method a Phosphorylation step is mentioned as well a ligation before the treatment with DpnI. On other publications ( Methods Mol Biol. 2014;1116:73-87) there is no mention of such step and right after the 2nd PCR DpnI is added to the mix and used for the E. coli transformation.
At a first glace in made sense to have such steps included but the other article made me wonder. Is it actually needed? If not how does the "linearize" vector get ligated in the cells since the ends are not phosphorilated?
p.s. there is no mention of the need of phosphorilated primers
Thanks in advance for the clarification
cheers
Gabriele
Offline
#2 2014-10-15 13:48:36
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
Hey Gabriele,
I think the difference is that the exponential Megaprimer method leads to completely linear vector which needs to be blunt end ligated, while the more standard version of RF-Cloning gives you a circularized, albeit nicked, plasmid. The transformed cells will repair the nicks all on their own, but are just going to degrade the linear DNA; hence the phosphorylation/T4-ligase step. I'm just skimming through the 2014 article by Peleg and Unger again (doi: 10.1007/978-1-62703-764-8_6) and they are not using the exponential method, which is probably why you aren't seeing reference to a ligation step.
Hope this clarifies!
Good luck, and it would be awesome if you share your experience with the exponential method after you've given it a whirl.
-Steve
Offline
#3 2014-10-15 14:03:20
- Dungeonlord
- Member
- Registered: 2014-10-13
- Posts: 2
Re: Is phosphorilation needed?
Thank you very much for the very quick reply!
i will probably set up the experiment with both methods
2nd PCR with EMP--> phosphorilation--> ligation --> DpnI
2nd PCR --> DpnI
I will see if i get major differences.
Thanks again
Gabriele
Offline
#4 2015-01-17 14:33:28
- Sand
- Member
- Registered: 2014-07-25
- Posts: 11
Re: Is phosphorilation needed?
Hey Gabriele,
I think the difference is that the exponential Megaprimer method leads to completely linear vector which needs to be blunt end ligated, while the more standard version of RF-Cloning gives you a circularized, albeit nicked, plasmid. The transformed cells will repair the nicks all on their own, but are just going to degrade the linear DNA; hence the phosphorylation/T4-ligase step. I'm just skimming through the 2014 article by Peleg and Unger again (doi: 10.1007/978-1-62703-764-8_6) and they are not using the exponential method, which is probably why you aren't seeing reference to a ligation step.
Hope this clarifies!
Good luck, and it would be awesome if you share your experience with the exponential method after you've given it a whirl.
-Steve
I find this an interesting question.
But I find it hard to understand why the RF (traditional method) leads to a more plasmid type shaped product while the EMP method leads to "perfect" linearized products.
Why is this?
I do not see why one method keeps the plasmid shape and the other does not. Both are nicked plasmids, so why does the nicked plasmid stays a "plasmid" in the RF method, but not in the EMP method?
Last edited by Sand (2015-01-17 15:19:27)
Offline
#5 2015-01-17 15:51:38
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
Hi Sand,
You're not alone in misunderstanding how the rf-cloning method leads to a circularized product. Some mental gymnastics are involved.
It all has to do with the positioning of priming sites. Both the 3' and 5' end of a good mega-primer will be bound to it's target parental plasmid, with some non-complementary sequence in the middle. As you know, priming is initiated at the 3' end, and then, because the DNA polymerase does not have 5'-3' exonuclease activity, synthesis stops when the daughter strand bumps into the 5' end of the mega-primer. This happens on the complement strand as well, and two nicks are present when these newly synthesized products anneal — offset by the length of the mega-primer.
In the next round of PCR, if a mega-primer binds to a daughter product, the 3' end it finds itself at the extreme 5' of the template. Meaning there is nothing to synthesize in the 5'-3' direction, and this is why the 2° PCR of an RF-cloning project is non-exponential.
I hope this clarifies, but I can imagine it's still a bit confusing. The best I can do is promise that the logic is sound, and encourage you to draw the process out on paper until you see how the pieces all fit together. You've probably already looked at it, but it might be useful to check out the diagram in the Q & A again.
Take care,
-Steve
Offline
#6 2015-01-17 16:23:35
- Sand
- Member
- Registered: 2014-07-25
- Posts: 11
Re: Is phosphorilation needed?
Hi Sand,
You're not alone in misunderstanding how the rf-cloning method leads to a circularized product. Some mental gymnastics are involved.
It all has to do with the positioning of priming sites. Both the 3' and 5' end of a good mega-primer will be bound to it's target parental plasmid, with some non-complementary sequence in the middle. As you know, priming is initiated at the 3' end, and then, because the DNA polymerase does not have 5'-3' exonuclease activity, synthesis stops when the daughter strand bumps into the 5' end of the mega-primer. This happens on the complement strand as well, and two nicks are present when these newly synthesized products anneal — offset by the length of the mega-primer.
In the next round of PCR, if a mega-primer binds to a daughter product, the 3' end it finds itself at the extreme 5' of the template. Meaning there is nothing to synthesize in the 5'-3' direction, and this is why the 2° PCR of an RF-cloning project is non-exponential.
I hope this clarifies, but I can imagine it's still a bit confusing. The best I can do is promise that the logic is sound, and encourage you to draw the process out on paper until you see how the pieces all fit together. You've probably already looked at it, but it might be useful to check out the diagram in the Q & A again.
Take care,
-Steve
But I understand this.
This is not my problem. The problem is that I do not see why in the RF system you end up with "plasmid" DNA (circle) while in the EMP you end up with a linear piece of DNA...
When you denature your DNA to amplify it => you have a linear piece of DNA ... (or am I missing something here..)
I just do not see why during RF the DNA would stay circulated while in the EMP it will not.
The shape/form or where the primers bind, I do not see how this can attribute to staying as a circle form or a linear form.
Are you saying it stays as a circle because you have a non exponential cloning in the 2th PCR ? I do not see what this has to do with the "form" of the DNA?
I understand that the parental part stays as a plasmid (its not nicked) but the generated piece of DNA , by the PCR, is nicked, so this will linearize. So why would this form a plasmid (circle) later?
Offline
#7 2015-01-17 16:49:16
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
You are right: when the parental plasmid denatures, it stays a circle, but when the daughter plasmids denature they become linear. The reason the daughter products re-anneal into circles again is because the relative positions of the 5' and 3' ends of the sense and antisense strands are offset by the length of the mega-primer; this leads to a nicked, circular product.
With the EMP process, the newly synthesized sense and antisense strands are perfectly matched up, and the PCR is designed to work in a very canonical fashion. That's why blunt end ligation is necessary in EMP.
Maybe it will help to think about how the insert behaves during each method. During 'normal' RF-Cloning, the insert itself is not copied at any point during the 2° PCR; it is simply a passenger component of the mega-primer. In the EMP method, the insert is copied along with the rest of the plasmid.
Does that make any more sense?
-Steve
Offline
#8 2015-01-17 17:56:16
- Sand
- Member
- Registered: 2014-07-25
- Posts: 11
Re: Is phosphorilation needed?
You are right: when the parental plasmid denatures, it stays a circle, but when the daughter plasmids denature they become linear. The reason the daughter products re-anneal into circles again is because the relative positions of the 5' and 3' ends of the sense and antisense strands are offset by the length of the mega-primer; this leads to a nicked, circular product.
With the EMP process, the newly synthesized sense and antisense strands are perfectly matched up, and the PCR is designed to work in a very canonical fashion. That's why blunt end ligation is necessary in EMP.
Maybe it will help to think about how the insert behaves during each method. During 'normal' RF-Cloning, the insert itself is not copied at any point during the 2° PCR; it is simply a passenger component of the mega-primer. In the EMP method, the insert is copied along with the rest of the plasmid.
Does that make any more sense?
-Steve
So somehow, the fact that the "insert" (the megaprimer, except the "binding parts of the primer") is not copied a circle is formed. So it has to do with some sort of "spherical force" attributed by this megaprimer (insert)?
I see what you mean (I think), but find it hard to understand why this leads to a circle formation. I guess it has to do with some molecular forces, but the details are not clear to me.
BTW: if I understand you correctly, the insert itself (the megaprimer) is never copied/created during the second PCR and thus this is only formed during the first PCR. This makes the amount of starting material (of megaprimer) pretty important.
Offline
#9 2015-01-17 18:44:10
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
Ha, well... I think the term 'Spherical Force' may sound a bit more mystical than it really is. Here, I've drawn a picture:
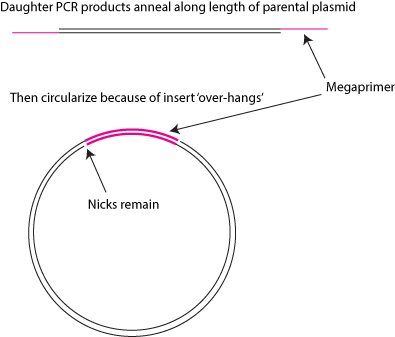
Does this help?
And yes, the amount of starting material is rather important. This is not your run-of-the-mill PCR reaction.
-Steve
Offline
#10 2015-01-17 18:56:37
- Sand
- Member
- Registered: 2014-07-25
- Posts: 11
Re: Is phosphorilation needed?
Ha, well... I think the term 'Spherical Force' may sound a bit more mystical than it really is. Here, I've drawn a picture:
http://www.rf-cloning.org/fluxbb/img/po … loning.png
Does this help?
And yes, the amount of starting material is rather important. This is not your run-of-the-mill PCR reaction.
-Steve
Ah yes, now I see it.
I should have drawn everything to see it visually!
Thanks.
Offline
#11 2015-01-17 19:00:46
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
My pleasure ![]()
Offline
#12 2015-01-17 19:25:26
- Sand
- Member
- Registered: 2014-07-25
- Posts: 11
Re: Is phosphorilation needed?
My pleasure
I wonder: if they do the EMP, why would they now use phosphorylated primers? It seems that this might do the trick to form a plasmid again!
A bacterium should be able to have a recircularization I think or perhaps this is a bit far fetched!
I wonder what if you used homologous overhangs on your primers when doing this: both ends would be homologous, I can imagine that it would be easier to have a recircularization then!
Offline
#13 2015-01-17 19:37:13
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
Nah, if you just transform bacteria with linear DNA, they chew it up. Wether you use pre-phosphorylated primers or T4 kinase, you will need to include a ligation step before transformation.
As far as using homologous overhangs on the primers, I'm not actually sure what you mean. If you add any extra sequence to the ends of the primers for EMP, it will be amplified along with everything else and you'll still be left with blunt ends.
Offline
#14 2015-01-17 20:05:20
- Sand
- Member
- Registered: 2014-07-25
- Posts: 11
Re: Is phosphorilation needed?
Nah, if you just transform bacteria with linear DNA, they chew it up. Wether you use pre-phosphorylated primers or T4 kinase, you will need to include a ligation step before transformation.
As far as using homologous overhangs on the primers, I'm not actually sure what you mean. If you add any extra sequence to the ends of the primers for EMP, it will be amplified along with everything else and you'll still be left with blunt ends.
Yes: blunt ends indeed, but they would be homologous to eachother!
What I was thinking is this: when you desing the two new primers (for the secondary PCR in EMP) you could make them so that both 5' ends are homologous to eachother. If they would be homologous for a few basepairs (I dunno 10 to 20 perhaps) would the bacterium not be able to link them using some sort of homologous end joining?
I am not sure you see what I mean. Perhaps I should try to draw it.
perhaps this will help:
5' NNNN ______________ NNNNN 3'
3' NNNN ______________NNNNN 5'
N-parts : are the parts that are homologous because you created the primers (for the second reaction) in such a way that they overlap that the 5' part (so after the PCR, you have this kind of DNA with the N-parts).
I see it more as some sort of "double strand break" in wich the bacterium thinks this is a double strand break and it will "recombinate" them again?
Is this clear?
Offline
#15 2015-01-17 21:12:37
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
I think I get what you're saying, and it may work... I worry that you're less likely to get the exact sequence you want though at the end of the day, because DS break repair/recombination can be a messy process. I'd say it might be a fun thing to play around with just to see what happens.
Give it a try ![]()
Offline
#16 2015-01-19 09:33:05
- Jan
- Member
- Registered: 2014-04-15
- Posts: 7
Re: Is phosphorilation needed?
I think I get what you're saying, and it may work... I worry that you're less likely to get the exact sequence you want though at the end of the day, because DS break repair/recombination can be a messy process. I'd say it might be a fun thing to play around with just to see what happens.
Give it a try
Fascinating discussion!
Check the link to the paper I included (its a free paper).
I am guessing this is what Sand means! I have not fully studied it, but it seems interesting.
Steve Bond wrote:Nah, if you just transform bacteria with linear DNA, they chew it up. Wether you use pre-phosphorylated primers or T4 kinase, you will need to include a ligation step before transformation.
As far as using homologous overhangs on the primers, I'm not actually sure what you mean. If you add any extra sequence to the ends of the primers for EMP, it will be amplified along with everything else and you'll still be left with blunt ends.Yes: blunt ends indeed, but they would be homologous to eachother!
What I was thinking is this: when you desing the two new primers (for the secondary PCR in EMP) you could make them so that both 5' ends are homologous to eachother. If they would be homologous for a few basepairs (I dunno 10 to 20 perhaps) would the bacterium not be able to link them using some sort of homologous end joining?
I am not sure you see what I mean. Perhaps I should try to draw it.perhaps this will help:
5' NNNN ______________ NNNNN 3'
3' NNNN ______________NNNNN 5'N-parts : are the parts that are homologous because you created the primers (for the second reaction) in such a way that they overlap that the 5' part (so after the PCR, you have this kind of DNA with the N-parts).
I see it more as some sort of "double strand break" in wich the bacterium thinks this is a double strand break and it will "recombinate" them again?
Is this clear?
Is this, perhaps not what you mean: http://www.sciencedirect.com/science/ar … 6114200161
I am not sure I understand you and the method (I just glanced at it, I was looking for further information because I want to try the RF-cloning in the near future.
So I stumbled upon this (above mentioned) paper!
Offline
#17 2015-01-19 13:39:22
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
Oh right! RAM cloning was discusses in a previous thread (here), and I had completely forgotten about it.
Skimming through the paper again, it does look like recombination leads to 20-30% weirdness, but it also seems to work where traditional RF may not. Very interesting.
-Steve
Offline
#18 2015-01-22 20:55:29
- Mitteke
- Member
- Registered: 2015-01-22
- Posts: 1
Re: Is phosphorilation needed?
Hi Sand,
You're not alone in misunderstanding how the rf-cloning method leads to a circularized product. Some mental gymnastics are involved.
It all has to do with the positioning of priming sites. Both the 3' and 5' end of a good mega-primer will be bound to it's target parental plasmid, with some non-complementary sequence in the middle. As you know, priming is initiated at the 3' end, and then, because the DNA polymerase does not have 5'-3' exonuclease activity, synthesis stops when the daughter strand bumps into the 5' end of the mega-primer. This happens on the complement strand as well, and two nicks are present when these newly synthesized products anneal — offset by the length of the mega-primer.
In the next round of PCR, if a mega-primer binds to a daughter product, the 3' end it finds itself at the extreme 5' of the template. Meaning there is nothing to synthesize in the 5'-3' direction, and this is why the 2° PCR of an RF-cloning project is non-exponential.
I hope this clarifies, but I can imagine it's still a bit confusing. The best I can do is promise that the logic is sound, and encourage you to draw the process out on paper until you see how the pieces all fit together. You've probably already looked at it, but it might be useful to check out the diagram in the Q & A again.
Take care,
-Steve
What I wonder: if you use EMP, you have to phosphorylate and ligate the fragments. Now I wonder: would you not get concatemers or pieces of DNA ligated completely wrong?
I am guessing this would lower the efficiency although it seems (according to the papers) not a problem. I have not found any information about this.
Offline
#19 2015-01-24 00:10:26
- Steve Bond
- Administrator
- Registered: 2014-01-23
- Posts: 130
Re: Is phosphorilation needed?
Hmmm, probably? I guess the solution is just to screen multiple colonies to ensure the predicted plasmid size is present, and of course sequence the insert as a final confirmation. I really don't have any experience with the EMP technique unfortunately...
Offline
Pages: 1